How to model immunity¶
In this tutorial, we are going to simulate the spread of Covid-19 in the case of a virus variant that is potentially resistant to immunity. We will first introduce the relevant immunity related variables that govern the model. Afterwards we will look at a simulation comparing how immunity and virus strains that are very resistant to it, interact in the model.
[1]:
%matplotlib inline
import warnings
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import sid
from sid.config import INDEX_NAMES
warnings.filterwarnings("ignore")
np.random.seed(0)
Preparation¶
For the simulation we need to prepare several objects which are identical to the ones from the tutorial on simulating multiple virus strain.
Initial states¶
[2]:
available_ages = [
"0-9",
"10-19",
"20-29",
"30-39",
"40-49",
"50-59",
"60-69",
"70-79",
"80-100",
]
ages = np.random.choice(available_ages, size=10_000)
regions = np.random.choice(["North", "South"], size=10_000)
initial_states = pd.DataFrame({"age_group": ages, "region": regions}).astype("category")
initial_states.head()
[2]:
| age_group | region | |
|---|---|---|
| 0 | 50-59 | North |
| 1 | 0-9 | South |
| 2 | 30-39 | South |
| 3 | 30-39 | North |
| 4 | 70-79 | South |
Contact Models¶
[3]:
def meet_distant(states, params, seed):
possible_nr_contacts = np.arange(10)
contacts = np.random.choice(possible_nr_contacts, size=len(states))
return pd.Series(contacts, index=states.index)
def meet_close(states, params, seed):
possible_nr_contacts = np.arange(5)
contacts = np.random.choice(possible_nr_contacts, size=len(states))
return pd.Series(contacts, index=states.index)
assort_by = ["age_group", "region"]
contact_models = {
"distant": {"model": meet_distant, "assort_by": assort_by, "is_recurrent": False},
"close": {"model": meet_close, "assort_by": assort_by, "is_recurrent": False},
}
Initial conditions¶
For the initial conditions, we assume a two-day burn-in period. On the first day, 50 people are infected with the base virus, on the second day one halve of 50 people has the old and the other halve the new variant.
Each column in the DataFrame is a categorical. Infected individuals have a code for the variant, all others have NaNs.
[4]:
infected_first_day = set(np.random.choice(10_000, size=50, replace=False))
first_day = pd.Series([pd.NA] * 10_000)
first_day.iloc[list(infected_first_day)] = "base"
infected_second_day_old_variant = set(
np.random.choice(
list(set(range(10_000)) - infected_first_day), size=25, replace=False
)
)
infected_second_day_new_variant = set(
np.random.choice(
list(set(range(10_000)) - infected_first_day - infected_second_day_old_variant),
size=25,
replace=False,
)
)
second_day = pd.Series([pd.NA] * 10_000)
second_day.iloc[list(infected_second_day_old_variant)] = "base"
second_day.iloc[list(infected_second_day_new_variant)] = "b117"
initial_infections = pd.DataFrame(
{
pd.Timestamp("2020-02-25"): pd.Categorical(
first_day, categories=["base", "b117"]
),
pd.Timestamp("2020-02-26"): pd.Categorical(
second_day, categories=["base", "b117"]
),
}
)
initial_conditions = {"initial_infections": initial_infections, "initial_immunity": 50}
Parameters¶
[5]:
epidemiological_parameters = pd.read_csv("infection_probs.csv", index_col=INDEX_NAMES)
assort_probs = pd.read_csv("assort_by_params.csv", index_col=INDEX_NAMES)
disease_params = sid.load_epidemiological_parameters()
params = pd.concat([disease_params, epidemiological_parameters, assort_probs])
[6]:
params
[6]:
| value | note | source | |||
|---|---|---|---|---|---|
| category | subcategory | name | |||
| health_system | icu_limit_relative | icu_limit_relative | 50.00 | NaN | NaN |
| cd_infectious_true | all | 1 | 0.39 | NaN | NaN |
| 2 | 0.35 | NaN | NaN | ||
| 3 | 0.22 | NaN | NaN | ||
| 5 | 0.04 | NaN | NaN | ||
| ... | ... | ... | ... | ... | ... |
| infection_prob | household | household | 0.20 | NaN | NaN |
| assortative_matching | close | age_group | 0.50 | NaN | NaN |
| region | 0.90 | NaN | NaN | ||
| distant | age_group | 0.50 | NaN | NaN | |
| region | 0.90 | NaN | NaN |
136 rows × 3 columns
Additional objects to model immunity¶
In the model, immunity is assumed to be a factor between 0 and 1. A non-zero immunity level can arise either through infection or vaccination. If an individual has contact with an infectious person state-dependent infection probability, say \(p\), is multiplied with the factor \((1 - \text{immunity})\). But we know that the effect of immunity is heterogeneous dependent on the virus strain. To model this phenomenon we include another virus dependent factor, which we call
immunity_resistance_factor. The updated infection probability is then
- :nbsphinx-math:`begin{align}
p leftarrow p times left(1 - text{immunity_resistance} times text{immunity} right) ,.
end{align}`
From this expression it is clear that, for the immunity resistance factor, values close to 0 imply that immunity barely affects the infection probability, while values close to one imply that the immunity level influences the infection probability directly. Now lets return to the simulation.
[11]:
for virus, cf, irf in [("base", 1, 1), ("b117", 1.3, 0.5)]:
params.loc[("virus_strain", virus, "contagiousness_factor"), "value"] = cf
params.loc[("virus_strain", virus, "immunity_resistance_factor"), "value"] = irf
Run the simulation¶
Now we will simulate this population over 200 periods. To compare the effects of the immunity resistance parameter we run the simulation for three parameter values: 0, 0.5 and 1.
[12]:
resistance_factors = [0, 0.5, 1]
df_dict = {}
for value in resistance_factors:
# update the immunity resistance factor
params.loc[("virus_strain", "b117", "immunity_resistance_factor"), "value"] = value
# simulate
simulate = sid.get_simulate_func(
initial_states=initial_states,
contact_models=contact_models,
params=params,
initial_conditions=initial_conditions,
duration={"start": "2020-02-27", "periods": 365},
virus_strains=["base", "b117"],
seed=2,
)
result = simulate(params=params)
df_dict[value] = result["time_series"].compute()
Start the simulation...
2021-02-25: 100%|██████████| 365/365 [00:44<00:00, 8.17it/s]
Start the simulation...
2021-02-25: 100%|██████████| 365/365 [00:37<00:00, 9.69it/s]
Start the simulation...
2021-02-25: 100%|██████████| 365/365 [00:35<00:00, 10.38it/s]
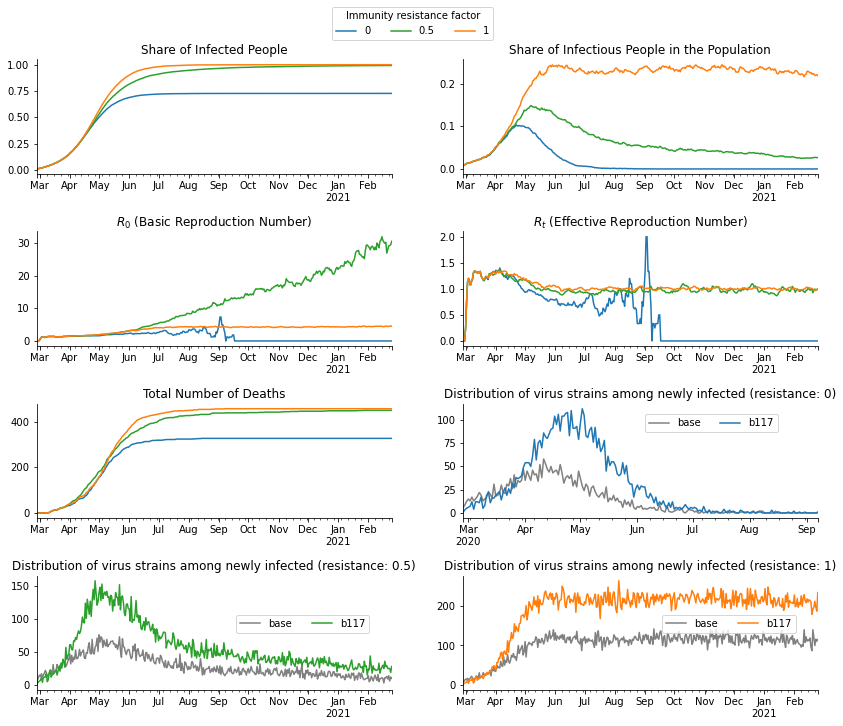
Let us take a look at various statistics of the sample.
[13]:
fig, axs = plt.subplots(4, 2, figsize=(14, 12))
fig.subplots_adjust(bottom=0.15, wspace=0.2, hspace=0.5)
axs = axs.flatten()
value_to_ax = {0: 5, 0.5: 6, 1: 7}
for value, color in [(0, "tab:blue"), (0.5, "tab:green"), (1, "tab:orange")]:
df_dict[value].resample("D", on="date")["ever_infected"].mean().plot(
ax=axs[0], color=color, label=value
)
df_dict[value].resample("D", on="date")["infectious"].mean().plot(
ax=axs[1], color=color
)
df_dict[value].resample("D", on="date")["dead"].sum().plot(ax=axs[4], color=color)
r_zero = sid.statistics.calculate_r_zero(df_dict[value], window_length=7)
r_zero.plot(ax=axs[2], color=color)
r_effective = sid.statistics.calculate_r_effective(df_dict[value], window_length=7)
r_effective.plot(ax=axs[3], color=color)
_df = (
df_dict[value]
.query("newly_infected")
.groupby([pd.Grouper(key="date", freq="D"), "virus_strain"])["newly_infected"]
.count()
.unstack()
)
_df["base"].plot(ax=axs[value_to_ax[value]], color="grey")
_df["b117"].plot(ax=axs[value_to_ax[value]], color=color)
for ax in axs:
ax.set_xlabel("")
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
axs[0].legend(
ncol=3,
bbox_to_anchor=(1.3, 1.5),
title="Immunity resistance factor",
)
axs[5].legend(loc="upper center", bbox_to_anchor=(0.70, 0.95), ncol=2)
axs[6].legend(loc="upper center", bbox_to_anchor=(0.75, 0.70), ncol=2)
axs[7].legend(loc="upper center", bbox_to_anchor=(0.75, 0.70), ncol=2)
axs[0].set_title("Share of Infected People")
axs[1].set_title("Share of Infectious People in the Population")
axs[2].set_title("$R_0$ (Basic Reproduction Number)")
axs[3].set_title("$R_t$ (Effective Reproduction Number)")
axs[4].set_title("Total Number of Deaths")
axs[5].set_title("Distribution of virus strains among newly infected (resistance: 0)")
axs[6].set_title("Distribution of virus strains among newly infected (resistance: 0.5)")
axs[7].set_title("Distribution of virus strains among newly infected (resistance: 1)")
plt.show()