How to simulate¶
This tutorial helps you to get started with sid by building a simple model. First, it focuses on the core objects which are needed for simulation. Then, the spread of the disease in the artificial population is simulated.
Are we all set? Let’s go!
[1]:
%matplotlib inline
import warnings
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import sid
from sid.config import INDEX_NAMES
warnings.filterwarnings(
"ignore", message="indexing past lexsort depth may impact performance."
)
The core objects¶
This section explains the core objects necessary to run a simulation with sid.
The initial_states¶
Since sid is an agent-based simulator, it needs information on agents or individuals in the model. This information is captured in the initial_states. The initial_states are a pandas.DataFrame where rows represent individuals and columns are characteristics, for example, age, geographic information, etc..
In this context, a state is a single row in the data frame representing an individual at a certain point in time. The characteristics contain all the information needed for the individual to make decisions such as going to work, demanding a test, or for policies to target the individual.
The following states include information on the age of individuals who fall into different age brackets. The age brackets are necessary for some stochastic processes in the model. For example, the probability of staying asymptomatic after an infection is age-dependent since elderly are more likely to experience strong symptoms.
The states also include very broad geographic information of the individuals which only there to speed up simulation.
Note
To give a little bit of background information on the matching algorithm: Later, the tutorial will allow for meetings between two randomly selected people in the model. Instead of searching for a contact partner among all people, it is easier to, first, sample a subset where the partner is in, such as a geographic cluster, and, after that, sample the specific individual.
Once there is an detailed information on the matching algorithm, it will be referenced here. Until then, please consult the API documentation on calculate_infections_by_contacts() and related functions.
We start with 10,000 individuals. Note that all characteristics are converted to categoricals which ensures that values are self-explanatory and save memory.
[2]:
n_individuals = 10_000
available_ages = [
"0-9",
"10-19",
"20-29",
"30-39",
"40-49",
"50-59",
"60-69",
"70-79",
"80-100",
]
ages = np.random.choice(available_ages, size=n_individuals)
regions = np.random.choice(["North", "East", "South", "West"], size=n_individuals)
initial_states = pd.DataFrame({"age_group": ages, "region": regions}).astype("category")
initial_states.head(5)
[2]:
| age_group | region | |
|---|---|---|
| 0 | 10-19 | East |
| 1 | 70-79 | West |
| 2 | 10-19 | South |
| 3 | 10-19 | North |
| 4 | 40-49 | East |
See also
You can read more about the states in the reference guides.
The contact_models¶
The contact models are key to how contacts between individuals can be modeled with sid. There are two types of contact models, random and recurrent models:
Recurrent contact models are used when people belong to a certain group and if they meet this group, they meet all participating members. You would use recurrent models to model school classes, households, and colleagues/work groups.
Random contact models are used when people do not always meet the same predefined groups, but contacts happen more randomly. Usually, people are more likely to meet a person from their own group, but there is also a lower probability of meeting someone from another group. Groups can be formed by combining any number of characteristics. If we would use age groups, we would say contacts are assortative by age. Each person also has a budget for the number of people she can meet. You would use random contact models, for example, to model meetings during freetime activities, infrequent contacts at work, or acquaintances.
In the following, we define a random contact model which lets each individual in the population meet 0-9 other people each day. The contacts are assortative by age and region since we want to make contacts more likely within the same regions and in the same age groups.
[3]:
def meet_people(states, params, seed):
np.random.seed(seed)
contacts = np.random.choice(np.arange(10), size=len(states))
return pd.Series(contacts, index=states.index)
[4]:
contact_models = {
"meet_people": {
"model": meet_people,
"assort_by": ["age_group", "region"],
"is_recurrent": False,
}
}
Contact models are stored as a dictionary. Our contact model is called "meet_people" and we define it in another dictionary. The dictionary has the following components.
"model"holds the function which maps individual characteristics with parameters to a number of contacts which can be an integer or a float – internally the number of contacts is rounded over the whole population while preserving the sum of contacts."assort_by"specifies the variables by which people are more likely to meet each other."is_recurrent"is an indicator for a whether the models represents recurrent or random contacts.
See also
You can read more about contact_models in the references guides.
The params¶
The parameters are a pandas.DataFrame and include medical and other relevant information. sid offers a prepared database which can be loaded and easily adjusted. The following code can be used for loading.
[5]:
params = sid.load_epidemiological_parameters()
params.head(5)
[5]:
| value | |||
|---|---|---|---|
| category | subcategory | name | |
| health_system | icu_limit_relative | icu_limit_relative | 50.00 |
| cd_infectious_true | all | 1 | 0.39 |
| 2 | 0.35 | ||
| 3 | 0.22 | ||
| 5 | 0.04 |
The index is a three dimensional multi index which allows to organize parameters hierarchically. If lower levels are not necessary, it is common to repeat the previous level.
For our model, we need to specify some additional parameters. First, we add the infection probability for our contact model. If a contact happens between an infectious and a susceptible person, the susceptible person becomes infected according to this probability.
[6]:
params.loc[("infection_prob", "meet_people", "meet_people"), "value"] = 0.05
Similarly, we specify for each contact model how the strength of the assortative matching.
We assume that 90% of contacts are with people from the same region and 50% with contacts from the same age group. It means that the probability of meeting someone from the same age group and region is 0.45. The rest of the probability mass will be split equally between the remaining groups formed by regions and age groups.
[7]:
params.loc[("assortative_matching", "meet_people", "age_group"), "value"] = 0.5
params.loc[("assortative_matching", "meet_people", "region"), "value"] = 0.9
We further have to specify parameters governing the immunity level and its waning. For a detailed explaination of these parameters see the tutorial notebook on how to model immunity.
[8]:
immunity_params = pd.read_csv("immunity_params.csv", index_col=INDEX_NAMES)
params = pd.concat((params, immunity_params))
See also
You can read more about params in the references guides.
The initial_conditions¶
Finally, we need to set the initial_conditions which describes the progression of the disease at the start of the simulation like who is infected or already immune, etc.. sid gives you the ability to start the progression of the infectious disease from patient zero. It also offers a simple interface to come up with complex patterns like individuals in all stages of the disease.
For now, we assume that there are 100 infected individuals and 50 with pre-existing immunity.
[9]:
initial_conditions = {"initial_infections": 100, "initial_immunity": 50}
See also
More information on the initial_conditions can be found in a dedicated how-to guide.
The simulation¶
Now, we are going to simulate this population for 200 periods. For that, we define a duration which is a dictionary with a "start" and a "periods" key or a "start" and "end" keys.
[10]:
duration = {"start": "2020-02-27", "periods": 200}
sid differentiates between the setup of the simulation and the actual simulation. That is why we need to build the simulation() function first.
[11]:
simulate = sid.get_simulate_func(
initial_states=initial_states,
contact_models=contact_models,
params=params,
initial_conditions=initial_conditions,
duration=duration,
seed=0,
)
After that, the function to simulate the data only depends on the parameters. Now, let us simulate our model!
[12]:
result = simulate(params=params)
Start the simulation...
2020-09-13: 100%|██████████| 200/200 [00:28<00:00, 7.07it/s]
The result object is a dictionary with two keys:
"time_series"is adask.dataframewhich holds the data generated by the simulation. Since it is not feasible to hold all the data from the simulation in memory, each day is stored on disk in compressed.parquetfiles.daskprovides a convenient interface to load all files and perform calculation on the data. If you want to work withpandas.DataFrames instead, convert thedask.dataframetopandaswithtime_series.compute()."last_states"holds adask.dataframeof the lastly simulated states including all data columns which are usually dropped to save memory. Thelast_statescan be used to resume a simulation.
[13]:
result.keys()
[13]:
dict_keys(['time_series', 'last_states'])
Here is a preview for each of the two objects.
[14]:
result["time_series"].head()
[14]:
| symptomatic | is_tested_positive_by_rapid_test | infectious | age_group | immunity | newly_vaccinated | cd_infectious_false | newly_infected | ever_infected | date | virus_strain | ever_vaccinated | knows_immune | newly_deceased | region | knows_infectious | needs_icu | new_known_case | n_has_infected | dead | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | False | False | False | 10-19 | 0.0 | False | -10001 | False | False | 2020-02-27 | NaN | False | False | False | East | False | False | False | 0 | False |
| 1 | False | False | False | 70-79 | 0.0 | False | -10001 | False | False | 2020-02-27 | NaN | False | False | False | West | False | False | False | 0 | False |
| 2 | False | False | False | 10-19 | 0.0 | False | -10001 | False | False | 2020-02-27 | NaN | False | False | False | South | False | False | False | 0 | False |
| 3 | False | False | False | 10-19 | 0.0 | False | -10001 | False | False | 2020-02-27 | NaN | False | False | False | North | False | False | False | 0 | False |
| 4 | False | False | False | 40-49 | 0.0 | False | -10001 | False | False | 2020-02-27 | NaN | False | False | False | East | False | False | False | 0 | False |
[15]:
result["last_states"].head()
[15]:
| age_group | region | ever_infected | infectious | symptomatic | needs_icu | dead | pending_test | received_test_result | knows_immune | ... | cd_knows_infectious_false_draws | cd_needs_icu_false_draws | cd_needs_icu_true_draws | cd_received_test_result_true_draws | cd_symptoms_false_draws | cd_symptoms_true_draws | group_codes_meet_people | date | period | n_contacts_meet_people | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10-19 | East | True | False | False | False | False | False | False | False | ... | -1 | -1 | -1 | 2 | 27 | -1 | 4 | 2020-09-13 | 621 | 5 |
| 1 | 70-79 | West | False | False | False | False | False | False | False | False | ... | -1 | -1 | 10 | 4 | 18 | 1 | 31 | 2020-09-13 | 621 | 3 |
| 2 | 10-19 | South | False | False | False | False | False | False | False | False | ... | -1 | 15 | -1 | 2 | 18 | -1 | 6 | 2020-09-13 | 621 | 1 |
| 3 | 10-19 | North | False | False | False | False | False | False | False | False | ... | -1 | -1 | -1 | 1 | 27 | -1 | 5 | 2020-09-13 | 621 | 0 |
| 4 | 40-49 | East | False | False | False | False | False | False | False | False | ... | -1 | 25 | -1 | 1 | 27 | 1 | 16 | 2020-09-13 | 621 | 6 |
5 rows × 49 columns
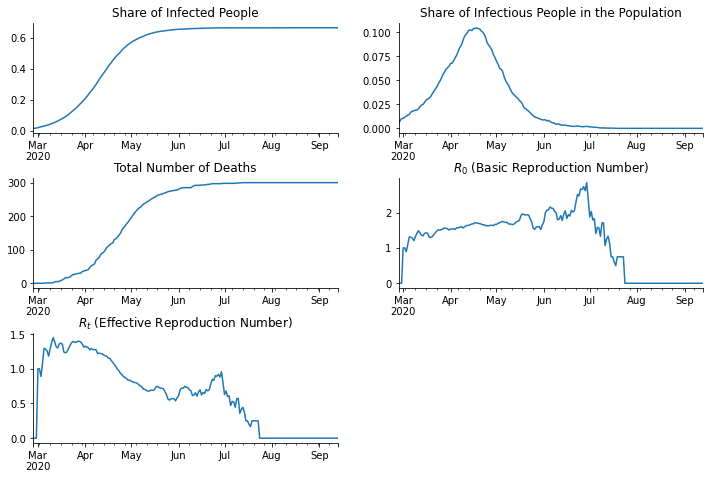
Let us take a look at various statistics of the sample.
[16]:
df = result["time_series"].compute()
[17]:
fig, axs = plt.subplots(3, 2, figsize=(12, 8))
fig.subplots_adjust(bottom=0.15, wspace=0.2, hspace=0.4)
axs = axs.flatten()
df.resample("D", on="date")["ever_infected"].mean().plot(ax=axs[0])
df.resample("D", on="date")["infectious"].mean().plot(ax=axs[1])
df.resample("D", on="date")["dead"].sum().plot(ax=axs[2])
r_zero = sid.statistics.calculate_r_zero(df, window_length=7)
r_zero.plot(ax=axs[3])
r_effective = sid.statistics.calculate_r_effective(df, window_length=7)
r_effective.plot(ax=axs[4])
for ax in axs:
ax.set_xlabel("")
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
axs[0].set_title("Share of Infected People")
axs[1].set_title("Share of Infectious People in the Population")
axs[2].set_title("Total Number of Deaths")
axs[3].set_title("$R_0$ (Basic Reproduction Number)")
axs[4].set_title("$R_t$ (Effective Reproduction Number)")
axs[5].set_visible(False)
plt.show()